Musculoskeletal Biology and Pathology
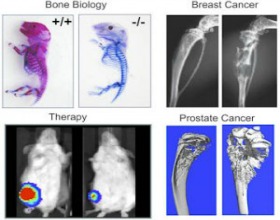 A main focus of our research for several decades has been exploration of molecular mechanisms that regulate skeletal development and remodeling. These studies center on commitment of mesenchymal stem cells to the osteoblast lineage, and the subsequent growth and differentiation of pre-committed osteoblasts. Our lab has identified several unique parameters of transcriptional control that regulate phenotype-restricted gene expression in the context of nuclear architecture (see Nuclear Structure and Function). We identified nuclear matrix protein 2 (NMP2) as an osteoblast-restricted, nuclear matrix-associated, transcription factor. NMP2, identified by other labs as AML3 or Cbfa1 (and later designated Runx2), is essential for bone formation in vivo and is mutated in Cleidocranial Dysplasia, an autosomal bone disorder.
A main focus of our research for several decades has been exploration of molecular mechanisms that regulate skeletal development and remodeling. These studies center on commitment of mesenchymal stem cells to the osteoblast lineage, and the subsequent growth and differentiation of pre-committed osteoblasts. Our lab has identified several unique parameters of transcriptional control that regulate phenotype-restricted gene expression in the context of nuclear architecture (see Nuclear Structure and Function). We identified nuclear matrix protein 2 (NMP2) as an osteoblast-restricted, nuclear matrix-associated, transcription factor. NMP2, identified by other labs as AML3 or Cbfa1 (and later designated Runx2), is essential for bone formation in vivo and is mutated in Cleidocranial Dysplasia, an autosomal bone disorder.
We have since continued our quest to identify biological roles of Runx2 in osteoblast proliferation and differentiation and have demonstrated its central place in balancing growth with differentiation in osteoblasts. Using genomics, we have discovered many target genes for Runx2, and through proteomic approaches, we have identified several co-regulatory proteins that contribute to Runx2-mediated gene expression. Of direct clinical relevance is our discovery that Runx2 is highly expressed in breast and prostate cancer cells metastasizing to bone. Exploration of these new dimensions to Runx-mediated control of gene expression represents a major focus of our research group.
We have also established the essential role of non-coding RNAs in the bone-lineage using a conditional Dicer null mouse model which has a high bone mass phenotype. We have examined the biological function of microRNAs at multiple stages during differentiation in various mesenchymal lineages (e.g., osteoblasts, chondrocytes and myoblasts), as well as identified a network of microRNAs that controls the osteoblast-related master regulator Runx2 (see Genetic and Epigenetic Regulation of Gene Expression). We are investigating the contribution of long non-coding RNAs to epigenetic control of osteoblast phenotype commitment. Genomic targets are being identified by ChIP-Seq. Functional consequences of conditional activation and suppression of long non-coding RNAs are being explored by CRISPR and antisense approaches.
Landmark Papers
Gordon JAR, Tye CE, Banerjee B, Ghule PN, van Wijnen AJ, Kabala FS, Page NA, Falcone MM, Stein JL, Stein GS, Lian JB. LINC01638 sustains human mesenchymal stem cell self-renewal and competency for osteogenic cell fate. Sci Rep. 2023 Nov 20;13(1):20314. doi: 10.1038/s41598-023-46202-z. PMID: 37985890; PMCID: PMC10662126.
Tye CE, Ghule PN, Gordon JAR, Kabala FS, Page NA, Falcone MM, Tracy KM, van Wijnen AJ, Stein JL, Lian JB, Stein GS. LncMIR181A1HG is a novel chromatin-bound epigenetic suppressor of early stage osteogenic lineage commitment. Sci Rep. 2022 May 11;12(1):7770. doi: 10.1038/s41598-022-11814-4. PMID: 35546168; PMCID: PMC9095685.
Bighetti-Trevisan RL, Almeida LO, Castro-Raucci LMS, Gordon JAR, Tye CE, Stein GS, Lian JB, Stein JL, Rosa AL, Beloti MM. Titanium with nanotopography attenuates the osteoclast-induced disruption of osteoblast differentiation by regulating histone methylation. Mater Sci Eng C Mater Biol Appl. 2021 Nov 13:112548. doi: 10.1016/j.msec.2021.112548. Epub ahead of print. PMID: 35012895.
Che X, Park NR, Jin X, Jung YK, Han MS, Park CY, Chun JS, Kim SG, Jin J, Kim HJ, Lian JB, Stein JL, Stein GS, Choi JY. Hypoxia-inducible factor 2α is a novel inhibitor of chondrocyte maturation. J Cell Physiol. 2021 Oct;236(10):6963-6973. doi: 10.1002/jcp.30356. Epub 2021 Mar 21. PMID: 33748969; PMCID: PMC8662706.
Freitas GP, Lopes HB, Souza ATP, Gomes MPO, Quiles GK, Gordon J, Tye C, Stein JL, Stein GS, Lian JB, Beloti MM, Rosa AL. Mesenchymal stem cells overexpressing BMP-9 by CRISPR-Cas9 present high in vitro osteogenic potential and enhance in vivo bone formation. Gene Ther. 2021 Dec;28(12):748-759. doi: 10.1038/s41434-021-00248-8. Epub 2021 Mar 8. PMID: 33686254; PMCID: PMC8423866.
Paradise CR, Galvan ML, Kubrova E, Bowden S, Liu E, Carstens MF, Thaler R, Stein GS, van Wijnen AJ, Dudakovic A. The epigenetic reader Brd4 is required for osteoblast differentiation. J Cell Physiol. 2020 Jun;235(6):5293-5304. doi: 10.1002/jcp.29415. Epub 2019 Dec 23. PMID: 31868237; PMCID: PMC8128078.
Tye CE, Boyd JR, Page NA, Falcone MM, Stein JL, Stein GS, Lian JB. Regulation of osteogenesis by long noncoding RNAs: An epigenetic mechanism contributing to bone formation. Connect Tissue Res. 2018 Dec;59(sup1):35-41. doi: 10.1080/03008207.2017.1412432. PMID: 29745821; PMCID: PMC5965257.
Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS.
A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011 Jun 14;108(24):9863-8. doi: 10.1073/pnas.1018493108. Epub 2011 May 31. PMID: 21628588; PMCID: PMC3116419.